suppressPackageStartupMessages(library(tidyverse))
library(palmerpenguins)Specifically, these snippets enable me to:
Customise
quartooutputGenerate
ggplot2charts in the style that I useFormat tables in my preferred way
Access databases inside and outside of
shinySolve common niggles in package building
To demonstrate these code snippets, I’ll use the tidyverse packages with data from palmerpenguins.
Note: In each case below, you can copy the code by hovering over it and selecting the clipboard icon in the top-right of the code chunk.
1. Customise quarto output
Given my frequent use of Quarto notebooks, I’ve gravitated to these settings that work best for me.
However, you can easily tweak these settings to suit you, as each option has an accompanying ‘tab auto-complete’ feature.
---
title: "Add title here"
subtitle: "Add subtitle here"
author:
- Robin Penfold
date: today
format:
html:
anchor-sections: true
code-copy: hover
code-fold: true
code-link: true
code-overflow: wrap
code-summary: "</>"
code-tools: false
df-print: paged
embed-resources: true
float: true
footnotes-hover: true
highlight-style: pygments
lang: en-GB
linkcolor: "#63431c"
mainfont: "Arial"
table-of-contents: true
toc-depth: 4
toc-title: " "
title-block-banner: "#edd9c0"
title-block-banner-color: "#63431c"
title-block-categories: false
editor_options:
chunk_output_type: inline
---2. Generate ggplot2 charts in the style that I use
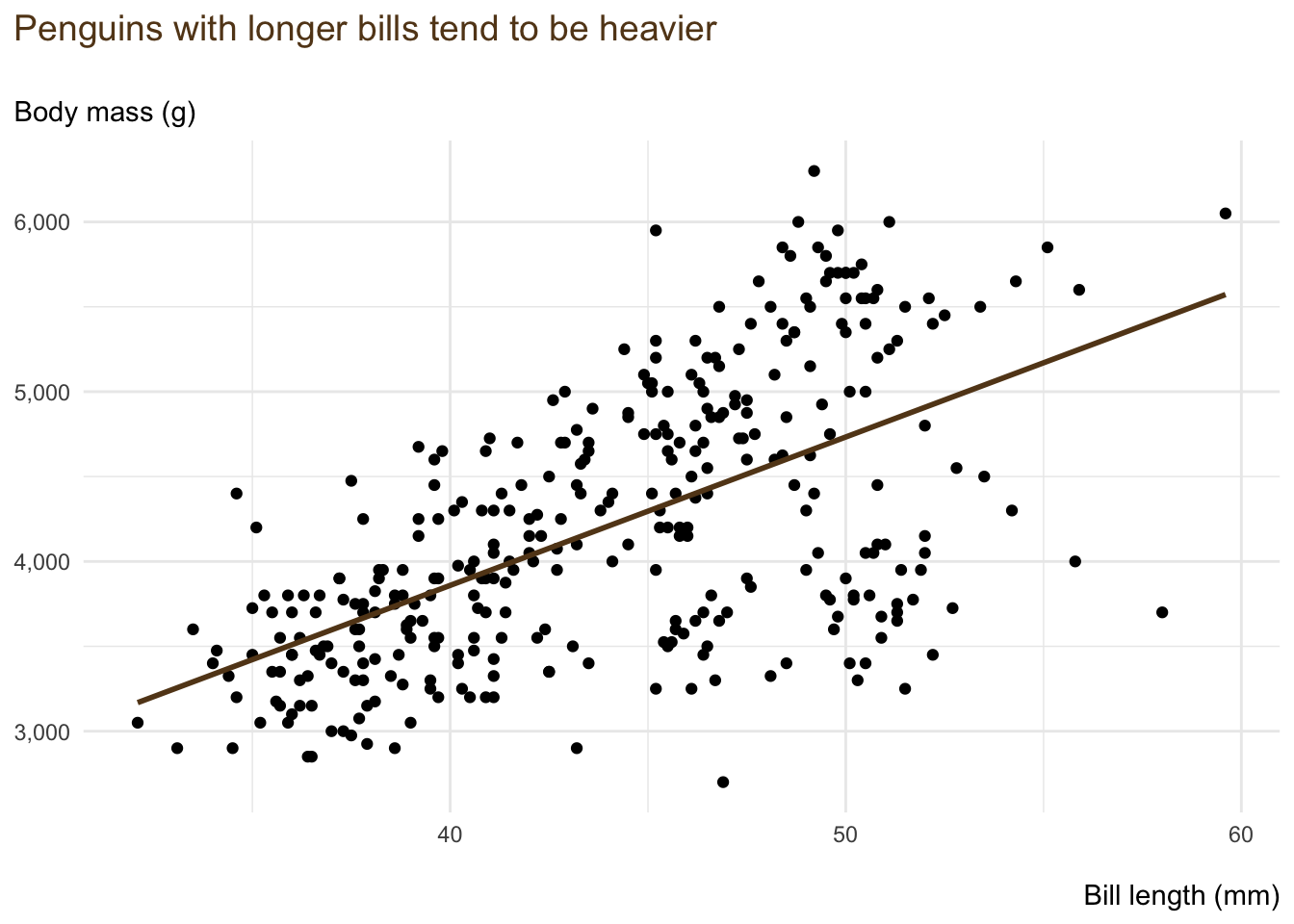
Over time, I have coalesced towards the following small chunk of code that builds (what I consider to be) a decent-looking chart in ggplot2.
penguins |>
ggplot(
aes(
x = bill_length_mm,
y = body_mass_g
)
) +
geom_point() +
geom_smooth(
method = "lm",
se = FALSE,
colour = "#63431c"
) +
labs(
title = "Penguins with longer bills tend to be heavier\n",
subtitle = "Body mass (g)",
x = "\nBill length (mm)",
y = NULL
) +
scale_y_continuous(labels = scales::label_comma()) +
theme_minimal() +
theme(
plot.title.position = "plot",
plot.title = element_text(size = 14, colour = "#63431c"),
axis.title.x = element_text(hjust = 1)
)
3. Format tables in my preferred way
I occasionally print tables using the out-of-the-box settings (admittedly tweaked by using the df-print: paged option in part 1 above). This generates a table as follows.
penguinsspecies <fct> | island <fct> | bill_length_mm <dbl> | bill_depth_mm <dbl> | flipper_length_mm <int> | |
|---|---|---|---|---|---|
| Adelie | Torgersen | 39.1 | 18.7 | 181 | |
| Adelie | Torgersen | 39.5 | 17.4 | 186 | |
| Adelie | Torgersen | 40.3 | 18.0 | 195 | |
| Adelie | Torgersen | NA | NA | NA | |
| Adelie | Torgersen | 36.7 | 19.3 | 193 | |
| Adelie | Torgersen | 39.3 | 20.6 | 190 | |
| Adelie | Torgersen | 38.9 | 17.8 | 181 | |
| Adelie | Torgersen | 39.2 | 19.6 | 195 | |
| Adelie | Torgersen | 34.1 | 18.1 | 193 | |
| Adelie | Torgersen | 42.0 | 20.2 | 190 |
Otherwise, I usually use reactable with the following tweaks (the results of which I omit for brevity) …
# library(reactable)
penguins |>
select(species, island, bill_length_mm, bill_depth_mm, body_mass_g) |>
reactable(
filterable = TRUE,
highlight = TRUE,
borderless = TRUE,
defaultPageSize = 5,
columns = list(
species = colDef(name = "Species", minWidth = 90, sticky = "left"),
island = colDef(name = "Island", minWidth = 90, sticky = "left"),
bill_length_mm = colDef(name = "Bill length", sticky = "right", filterable = FALSE, format = colFormat(separators = TRUE, digits = 1)),
bill_depth_mm = colDef(name = "Bill depth", sticky = "right", filterable = FALSE, format = colFormat(separators = TRUE, digits = 1)),
body_mass_g = colDef(name = "Body mass", sticky = "right", filterable = FALSE, format = colFormat(separators = TRUE, digits = 0))
)
)… or I use DT (i.e. the datatable package).
library(DT)
penguins |>
select("Species" = species, "Island" = island, "Bill length" = bill_length_mm, "Bill depth" = bill_depth_mm, "Body mass" = body_mass_g) |>
datatable(
rownames = FALSE,
width = "100%",
options=list(
dom = 'tip',
pageLength = 5
)
) |>
formatRound(
columns = 3:4,
digits = 1
) |>
formatRound(
columns = 5,
digits = 0
)4. Access databases inside and outside of shiny
I use the wonderful DBI and dbplyr all the time, not least for exploratory analysis.
(Note that whilst I typically don’t use SQLite, I will do so here, as it plays better with my website architecture.)
library(RSQLite)
con <- dbConnect(RSQLite::SQLite(), ":memory:")
dbWriteTable(con, "instruments", dplyr::band_instruments)
dbWriteTable(con, "members", dplyr::band_members)
dbListTables(con)[1] "instruments" "members" In this example, we create an object (con) for connecting to the SQLite database, where we add two tiny tables, called instruments and members. We can then explore these tables.
tbl(src = con, "instruments")name <chr> | plays <chr> | |||
|---|---|---|---|---|
| John | guitar | |||
| Paul | bass | |||
| Keith | guitar |
tbl(src = con, "members")name <chr> | band <chr> | |||
|---|---|---|---|---|
| Mick | Stones | |||
| John | Beatles | |||
| Paul | Beatles |
Even better, we can explore the tables when they are combined and tidied. (Whilst the code appears to return all the data, that’s only because our tables are uncommonly small.)
tbl(src = con, "instruments") |>
left_join(
tbl(src = con, "members"),
by = "name"
) |>
filter(band == "Beatles")name <chr> | plays <chr> | band <chr> | ||
|---|---|---|---|---|
| John | guitar | Beatles | ||
| Paul | bass | Beatles |
Once you have what you need, assign a name to the code and append it with |> collect().
Whilst this functionality is great outside of shiny, it is often more valuable within it. (After all, these apps can be a really safe and simple way for users to access a corporate database.)
To do so, some other tweaks are required within shiny’s server functionality, as illustrated below.
data_chosen <- shiny::reactive({
shiny::req(input$dataset)
main_data |>
dplyr::filter(name_dataset == input$dataset) |>
dplyr::mutate(id = as.integer(id))
})
data_chosen_id <- shiny::reactive(
quote({data_chosen()$id}),
quoted = TRUE
)
data_calculated <- shiny::reactive({
arbitrary_function(
con,
arbitrary_argument = data_chosen_id()
)
})5. Solve common niggles in package building
When I’m building packages, I often get dinged with notes or warnings about two common package niggles.
The first of these is non-ASCII characters. With apologies for the person who showed me, and who I now can’t recall, you can find these characters by:
Clicking
CTRL+Fin RStudioSelecting the Regex tick-box
Entering:
[\u0080-\uFFFF]or[^\x00-\x7F]as the search term
From there, you can use stringi::stri_escape_unicode('@') to get the Unicode equivalent for @. (Note that you also might need to remove the initial ‘\’ on Windows.)
The second niggle of package building concerns variable binding. It occurs during the package check and creates a note along the following lines.
no visible binding for global variable
‘ABC’In this case, we need to do something of the form below (from my simple package p0bservations that you can find here).
#' @title Calculate income net of UK tax and National Insurance
#'
#' @description This function ...
#' @param income_taxable The taxable income level ...
#' @param tax_year_end The calendar year in which the tax year ends ...
#' @export
#' @examples
#' \dontrun{
#' calc_income_net(income_taxable = 38000, tax_year_end = 2022L)
#' }
#'
#' @importFrom rlang .dataOnce we have added the line of #' @importFrom rlang .data, we can call the variables as follows (i.e. as before, but preceded by .data$).
year_tax_end_options <- p0bservations::tax_parameters |>
dplyr::distinct(.data$year_tax_end) |>
dplyr::pull(.data$year_tax_end)Once again, I hope that this aide-mémoire helps you as well as me!